Last modified 9/1/98.
Dr. LandryProtein-protein interactions are fundamental to all biological processes. An obvious example of importance to physiology and disease is the series of regulatory protein-protein interactions that mediate activation of an antigen presenting cell such as a macrophage:
All of these interactions are readily reversible; meaning, they
are characterized by only modest binding affinity. The dissociation
constants (Kd) are in the nanomolar to micromolar range. For comparison,
the Kd for dissociation of a multisubunit enzyme may be in the
picomolar to nanomolar range. The affinity is critically important
for the physiological process because it governs the rate of dissociation.
![]() ,
,
where koff is the off-rate of the ligand in sec**(-1) and kon
is the on-rate in (M*sec)**(-1).
Usually the on-rate of complex formation is controlled by the rate of diffusion. For protein-protein binding, kon might be as fast as 10**5 (M*sec)**(-1). Thus, for a Kd of 10**(-6) M (micromolar), the off-rate is (10**(-6))(10**5) or 0.1 sec**(-1). As you can see, if signal transduction must wait for dissociation of the upstream complex, then the signaling process may take a minute or so.
The same large opposing forces govern protein-protein interactions as govern protein folding. Favorable hydrophobic interactions in the contact interface are nearly balanced by the unfavorable entropy loss of restricting the relative motion of the two molecules. Intermolecular hydrogen bonds and electrostatic interactions are important for specficity. Typical protein-protein interfaces exhibit intermolecular contacts by an average of 15 amino acids on each protein. Nevertheless, in the case of growth hormone binding to its receptor and in antigen-antibody binding, individual replacement of only a certain few of these residues by alanine significantly diminishes binding [Jin, L., Cohen, F. E. & Wells, J. A. Structure from Function - Screening Structural Models with Functional Data. Proc. Natl. Acad. Sci. U. S. A. 91, 113-117 (1994); Clackson, T. & Wells, J. A. A hot spot of binding energy in a hormone-receptor interface. Science 267, 383-386 (1995)].
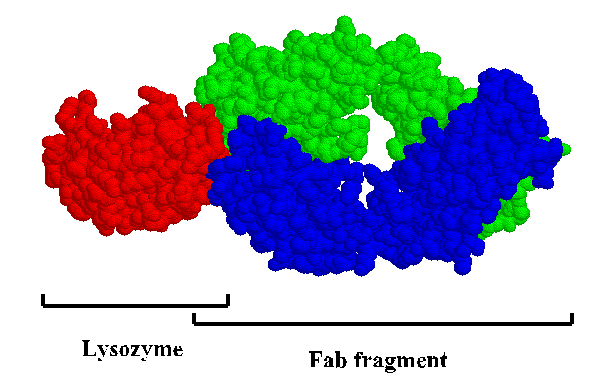
This is a "space-filled" rendering of a lysozyme-antibody complex. To get a close-up look at the interface, view the kinemage .
In response to many stresses, including heat, oxidizing conditions, and exposure to toxic compounds, all cells produce a common set heat shock proteins (Hsps). Experiments in E. coli, yeast, fruit flies and mice have shown that increased expression of these proteins can protect the organism against stress-induced damage. The classic phenomenon of acquired thermotolerance led to the identification of the major classes of heat shock proteins in E. coli. Cells given a non-lethal preshock at 42 oC subsequently survive an otherwise lethal exposure to 46 oC. The preshocked cells stopped synthesis of the normal spectrum of proteins and began to synthesize a few proteins at high level.
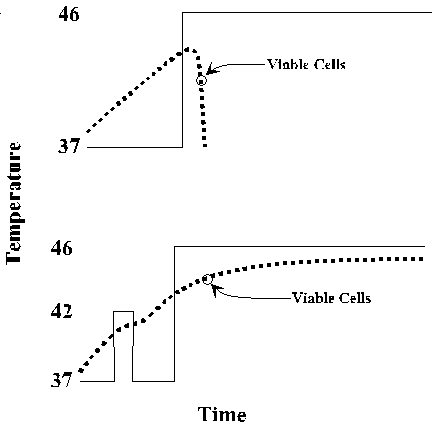
A very similar response occurs in all organisms. Increased expression of Hsps is mediated at multiple levels, mRNA synthesis, mRNA stability, and translation efficiency. Elevated synthesis of Hsps persists only through the initial period of stress. Even if the organism continues to be exposed to high temperature after the initial shock, Hsp expression drops, and Hsp levels return to normal. The heat shock response is distinct from adaptive responses that an organism may undergo when its environment changes gradually.
Most, but not all, heat shock proteins are molecular chaperones. Molecular chaperones bind and stabilize proteins at intermediate stages of folding, assembly, translocation across membranes and degradation. Heat shock proteins have been classified by molecular weight, for example, Hsp70 for the 70 kDa heat shock protein. Hsp70 null mutants of E. coli cannot grow at elevated temperature. Hsp100 null fruit flies lose their capacity for acquired thermotolerance. Heat shock proteins are among the most well-conserved proteins known. Amino acid sequences for Hsp70s of E. coli and man are almost 50% identical.
Hsp functions are required at normal temperatures, but the level
of expression is reduced, and the identity of the Hsp may be different.
In E. coli, Hsp70 is expressed at much reduced level in
unstressed conditions, but it plays an important role in export
of proteins to the periplasm and in protein degradation. In yeast,
there are 13 different Hsp70 proteins. The heat-induceable version
of yeast cytosolic Hsp70 is not expressed at normal temperatures,
but a constitutively expressed heat shock protein cognate (Hsc70)
participates in protein translocation across membranes and other
functions.
| Heat Shock Protein | Cohorts | Function, etc. |
| Hsp100 | ? | ATPse; dissociates aggregates, facilitates proteolysis; essential in yeast for acquired thermotolerance; essential for yeast prion propagation |
| Hsp90 | Immunophilins, Hsp70, others | stabilizes proteins prior to complete folding or activation; forms stable complexes with inactive glucocorticoid receptor and other transcription factors; most abundant non-ribosomal protein (cytosolic version); most abundant protein in endoplasmic reticulum (ER version) |
| Hsp70 | Hsp40 | ATPase; stabilizes proteins prior to complete folding, transport across membranes and proteolysis; found associated with misfolded and unassembled proteins, e.g., mutant p53 in cytosol or immunoglobulin heavy chains in ER of cells that don't make light chains; homologs in mitochondria and chloroplasts |
| Hsp60 | Hsp10 | ATPase; promotes efficient folding; only in mitochondria and chloroplasts of eukaryotes; distant homolog in cytosol is specialized for folding actin and tubulin; a.k.a., chaperonin |
| Hsp25 | ? | blocks aggregation; involved in regulation of actin assembly/disassembly |
All molecular chaperones recognize hydrophobic features of unfolded substrate polypeptides, thereby discriminating against folded proteins. Both Hsp70 and Hsp60 use ATP to control binding and dissociation of substrate polypeptides. Crystal structures of Hsp70 and Hsp60 reveal differences in mechanism.
Hsp70 proteins contain two domains, an amino-terminal ATPase domain and a carboxy-terminal peptide-binding domain. The secondary and tertiary structure of the ATPase domain is almost identical to that of actin. The Hsp70 peptide-binding domain binds a seven-residue peptide in an extended conformation between a beta-sheet subdomain and alpha-helical subdomain. Two or three hydrophobic residues of the peptide are buried in pockets of the beta subdomain, and peptide backbone NH and CO groups form hydrogen bonds with sidechain and backbone groups on the beta subdomain. The ends of the substrate peptide extend out from either side of the peptide-binding domain. The alpha domain clamps down loops of the beta subdomain, pinning the peptide in place. It is thought that ATP binding to the ATPase domain triggers substrate release by causing the alpha domain to bend upwards at a flexible junction near the middle of the long helix that extends over the peptide.
The peptide-binding domain of E. coli Hsp70 (DnaK).
The substrate peptide is shown in black. For a better look, view
the structure 1dkx.pdb using Rasmol. 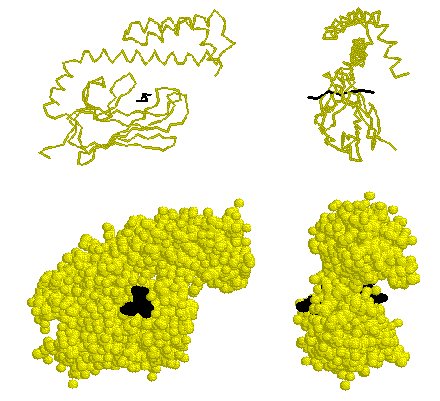
Hsp60 is somwhat more complicated because its oligomeric structure is a critical feature of its mechanism. Hsp60 proteins are composed of 14 subunits arranged in two stacked 7-membered rings. An unfolded protein binds to large hydrophobic surfaces inside the central cavity. ATP binding triggers rotation of the Hsp60 subunits such that the hydrophobic surface turns toward the neighboring Hsp60 subunit. This conformational change is coordinated around the ring by binding of Hsp10 which closes the cavity with the substrate inside. Perhaps this creates a chamber for the protein to fold without the possibility of aggregation with other unfolded proteins.
E. coli Hsp60 (GroEL) "top view."
C-alpha trace colored by crystrallographic B-factors. Red indicates
high mobility; blue low mobility. The inside of the apical domains,
where unfolded substrates are thought to bind, are the most mobile.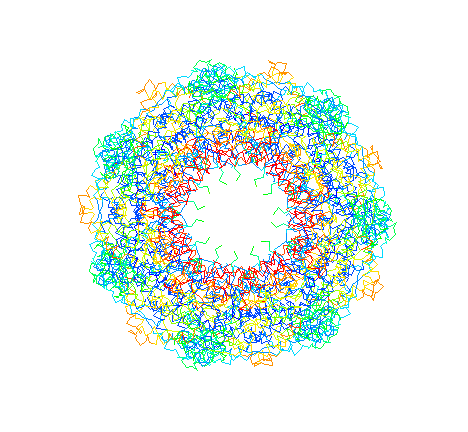
GroEL and GroES "side view." GroES binds via
mobile loops that extend down to contact the substrate-binding
domains of GroEL. Here is a link to more about mobile loops.
Despite the obvious importance of stress responses, only recently has scrutiny focused on the role of heat shock proteins in the control of disease pathology and in the survival and virulence of pathogens. Knowlege about Hsp functions in bacteria is much further advanced than in eukaryotes, but already some hints of Hsp involvement in mammalian diseases have emerged. Here is a list of phenomena.
End of document