Ph.D. (Duke, 1988)
1430 Tulane Ave., Box SL-43
New Orleans LA 70112
|
|
Ph.D. (Duke, 1988) |
| Phone: (504) 584-2453 | |
| FAX: (504) 584-2739 | |
| Address:
1430 Tulane Ave., Box SL-43 New Orleans LA 70112 | |
| Email: JNolan@tulane.edu |
Research Interests
The RNase P Holoenzyme. We are
examining structural aspects of RNA-protein interactions using the bacterial
ribonuclease P (RNase P) holoenzyme as a model system. RNase P consists
of a large RNA and a small protein subunit which together cleave 5'-precursor
sequences from tRNA transcripts. In vitro at high ionic strength, the RNA
subunit is the catalytic moiety and the protein is dispensable for catalysis.
Thus, RNase P is a member of an unusual class of enzyme known as ribozymes,
which use an RNA molecule, rather than a protein, as a catalyst. Although
the RNase P protein is not required for activity in vitro, it is essential
for viability in vivo. In addition, the kinetics of the holoenzyme reaction
in vitro differs dramatically from the RNA-alone reaction in salt requirement,
substrate specificity, and reaction rate. While much is known about the
RNA subunit of the enzyme, little is known about the mechanism by which
the protein moiety mediates its pleiotropic effects on the enzymatic reaction,
which are required for function in vivo.
We are using a detailed structural analysis of the RNase P RNA-protein
complex to investigate the how the protein binds to RNase P RNA, and how
this binding affects the reaction mechanism, including changes in how the
RNase P RNA interacts with its tRNA substrate in the presence of RNase
P protein. These problems are being studied by a variety of techniques.
The site of RNase P RNA with which the protein interacts is being studied
by chemical and enzymatic footprinting of the holoenzyme complex. We are
incorporating crosslinking reagents at specific sites of both the RNA and
protein subunits to further characterize the sites of interaction between
the two subunits. Regions of the RNase P protein important for interaction
with RNase P RNA will also be identified by in vitro and in vivo analyses
of mutations in the RNase P protein.
The results of these studies will shed light on the RNase P protein
and its function in the holoenzyme which, in turn, will enhance understanding
of RNA enzyme mechanism, and RNA-protein as well as RNA-RNA interactions.
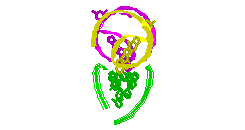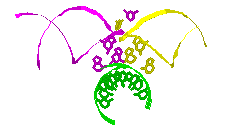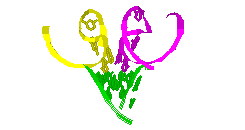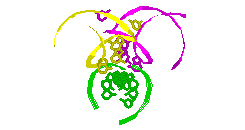 RNA Modeling. We
are also interested in modeling the structure of the RNase P RNA. This
is a primary research interest in the laboratory of Norm
Pace. I have modeled a tertiary structural element of RNase P RNA structure
using the MC-SYM
modeling program based upon phylogenetic comparative analysis done by Jim
Brown (see the abstract).
You can see the model
of this structure if you have the Chime
plug-in for viewing PDB files.
RNA Modeling. We
are also interested in modeling the structure of the RNase P RNA. This
is a primary research interest in the laboratory of Norm
Pace. I have modeled a tertiary structural element of RNase P RNA structure
using the MC-SYM
modeling program based upon phylogenetic comparative analysis done by Jim
Brown (see the abstract).
You can see the model
of this structure if you have the Chime
plug-in for viewing PDB files.
Publications
My Favorite Links
| Tulane Faculty Research | Tulane Biochemistry Home |