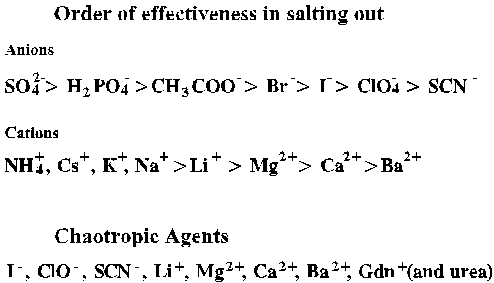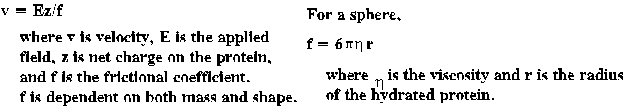Biochemistry 601
Protein Solubility, Purification, and Analysis
Last modified 8/23/99.
Dr. Landry
Rm. 6055
landry@mailhost.tcs.tulane.edu
Reading assignments:
Bibliography:
-
Voet, D. and Voet, J.G. (1990). Biochemistry, John Wiley and Sons,
New York.
Objectives
Become familiar with the following phenomena:
-
The ionic strength dependence of protein solubility depends on the
nature of the protein and on both the concentration and nature of the salt.
Differential solubility in ammonium sulfate solutions forms the basis of
an effective protein purification step.
-
The addition of salt can increase protein solubility (salting in)
or decrease solubility (salting out).
-
For a given ionic species the effectiveness of salting out parallels the
effectiveness of protein structure stabilization. The Hoffmeister series
rank ions on this basis.
-
A protein is minimally soluble near its isoelectric point (pI).
-
The rate of protein migration in non-denaturing electrophoresis depends
on the mass, shape and charge of the protein.
-
In SDS-PAGE, SDS and reducing agents equalize the charge/mass ratio
of proteins so that they are separated only on the basis of size (by gel
filtration).
-
Ion exchange chromatography separates proteins on the basis of electrostatic
interactions that depend on the protein's charge density, the concentration
of ionic species in solution, and the pH.
-
Gel filtration chromatography is both a purification technique and
analytical tool that separates macromolecules on the basis of size and
shape.
Protein Solubility
Salt Effects
Ionic strength may be expressed as,
![I=1/2[c(1)z(1)**2+c(2)z(2)**2....+c(i)z(i)**2]](ionic.gif)
-
where c(i) and z(i) are the concentration and charge of ionic
species i.
-
Salting in
-
Solubility at low ionic strength increases with salt concentration. Pairing
of salt ions with charged groups of the protein shields intramolecular
repulsion. Precipitates tend to be irreversible, suggesting that the protein
is denatured.
-
Salting out
-
Solubility at high ionic strength decreases with salt concentration. Solvent
activity toward solubilization of hydrophobic solutes is reduced. When
used for the purpose of salting out, solution additives are called precipitants.
-
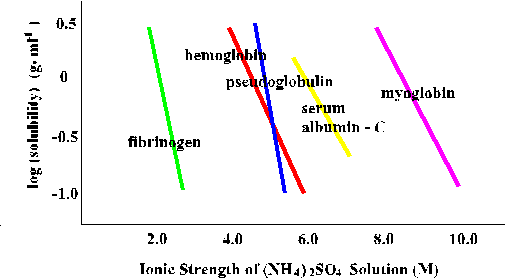
Ammonium Sulfate Fractionation
-
At a given ammonium sulfate concentration, one protein can be very soluble
while another is essentially insoluble.
-
-
Hoffmeister Series
-
Salt ions pair with ionized groups on proteins with varying effects on
protein stability and solubilization. The most effective protein precipitants
also are the most effective stabilizers. Thus, proteins may be stored in
a highly stable form as the ammonium sulfate precipitate. Many enzymes
are packaged and sold in this state. Good precipitants reduce the activity
of the solution toward solubilization of polypeptides, exactly opposite
the behavior of chaotropes.
-
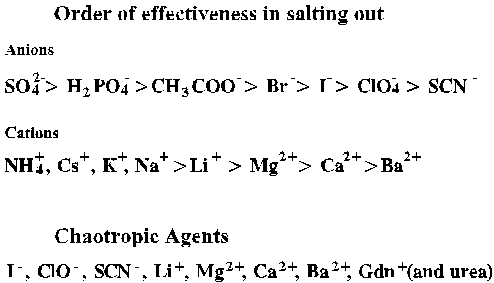
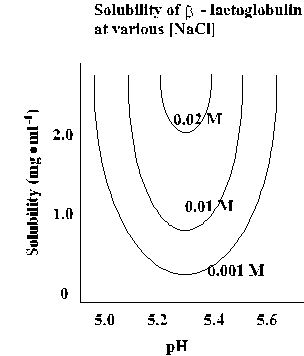
Effect of pH
Protein solubility is at a minimum near the isoelectric point (pI) of the
protein.
Polyacrylamide Gel Electrophoresis
Polyacrylamide gel electrophoresis (PAGE) has become the method of choice
for the analytical separation of proteins and low molecular weight nucleic
acids.
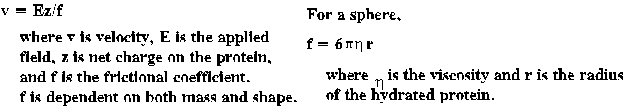
Owing to their uniform shape and charge/mass ratio, DNA molecules are
easily separated on the basis of size by gel filtration (or gel permeation),
and their rate of migration is inversely related to the log of the molecular
weight. RNA molecules typically have more variable shapes due to variable
secondary structure, but these structures can be disrupted by chaotropic
agents such as strong base. Proteins are neither uniform in shape nor uniform
in charge/mass ratio. While non-denaturing PAGE effectively resolves protein
mixtures, the relationship of size to rate of migration is not the same
for different proteins. One can estimate the size of the protein by running
unknown and control samples on a series of gels prepared with increasing
polyacrylamide concentrations and then plotting the change in migration
of the unknown and controls against polyacrylamide concentration. This
method eliminates the influence of charge/mass ratio. However, rigorous
analysis of protein native molecular weight is more commonly carried out
by gel filtration chromatography or analytical ultracentrifugation.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)
is a versatile and powerful tool for analyzing the polypeptide composition
of protein samples, and it can provide an accurate estimate of the subunit
molecular weight. SDS molecules form micelles, and each polypeptide is
incorporated into a micelle by interacting hydrophobically with the detergent's
alkyl tails. Variability in polypeptide shape and charge/mass ratio are
eliminated by the uniform shape of the micelles and by the essentially
constant ratio of 3 SDS molecules per amino acid residue. The large number
of sulfate ions in the micelle overwhelms the effect of charges on the
polypeptide itself.
Reagents for Protein Analysis
-
N,N'-methylenebisacrylamide
-
polyacrylamide crosslinker; Stryer, p. 46
-
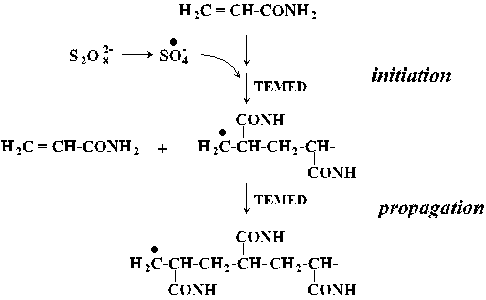
N,N,N',N'-tetramethylethylenediamine (TEMED)
-
radical stabilizer
-
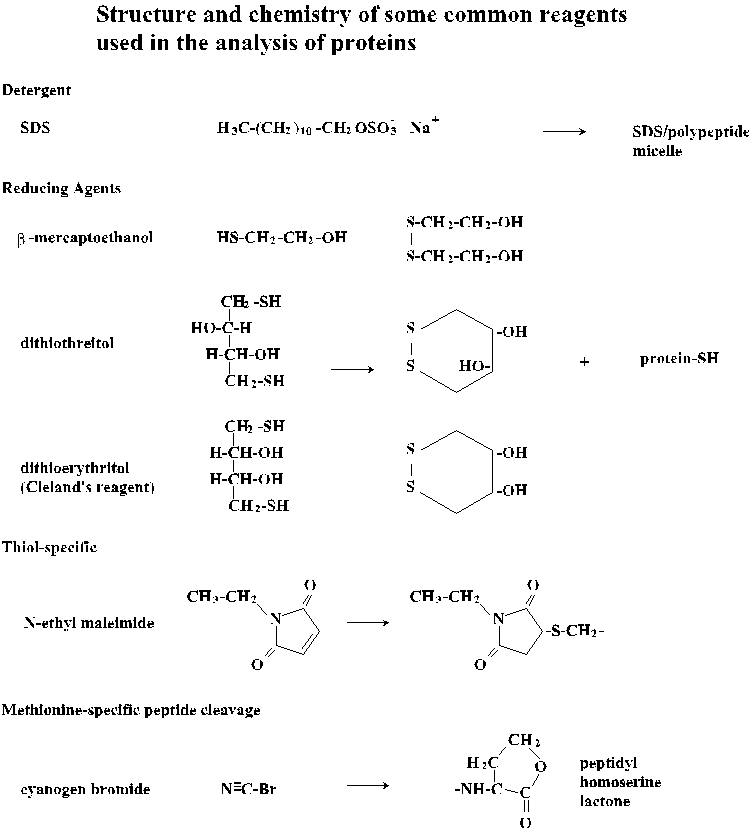
sodium dodecyl sulfate (SDS)
-
formation of detergent/protein micelles; Stryer, p. 47
-
dithiothreitol (DTT), dithioerythritol (DTE), 2-mecaptoethanol
-
reduction of disulfide bonds; Stryer, p. 57
-
phenyl isothiocyanate (PITC)
-
Edman degradation; Stryer, p. 55
-
cyanogen bromide (CNBr)
-
cleavage of methionine amino acyl peptide bonds; Stryer, p. 56
-
trypsin
-
cleavage of lysine and arginine amino acyl peptide bonds; Stryer p. 56
-
iodoacetate
-
modification of cysteine (also reactive to Met, His, Asp, Glu, Lys); Stryer,
p. 57
-
performic acid
-
modification of cystine (also reactive to Cys, Met); Stryer, p. 58
![I=1/2[c(1)z(1)**2+c(2)z(2)**2....+c(i)z(i)**2]](ionic.gif)
![I=1/2[c(1)z(1)**2+c(2)z(2)**2....+c(i)z(i)**2]](ionic.gif)