Hammerhead Ribozyme
References:
1. Pley, H.W., Flaherty, K.M. and McKay, D.B. (1994) Three-Dimensional
Structure of a Hammerhead Ribozyme. Nature 372, 68-74.
[Medline Abstract]
2. Scott, W. G., Finch, J. T. and Klug, A. (1995). The Crystal
Structure of an All-RNA Hammerhead Ribozyme: A Proposed Mechanism
for Catalytic Cleavage. Cell. 81, 991-1002. [Medline Abstract]
I. Ribozyme and Substrate

The minimal consensus structure for the hammerhead ribozyme
is shown above. The sequences shown as outlines are required for
efficient cleavage. Residues indicated by N can be any
base and N´ indicates the complement of the base
with which it is paired. The H at the cleavage site means
any base but G. The lines at the ends of stems I, II
, and III indicate that any number of bases may connect
the two halves of each stem, or they need not be connected at
all. In the 3-D structure to the left and the secondary structure
above, the strand cleaved is shown in red
and the rest of the molecule is shown in blue.
II. Tertiary structure
In the secondary structure below, the hammerhead is colored
as it is in reference 1. Click here <

An alternative representation can be seen using this button
<
IIa. U-turn
To see the atoms oriented as in Fig. 3 of reference 1,
click here<>. This is the
region of the active site and the U-turn
motif. You can show the H-bonds which help hold the U-turn
in place and their distances by clicking here <
>. You can also highlight the H-bonds of the regular Watson-Crick
pairs in this region by clicking here <
>. You can toggle off the H-bonds by reclicking the buttons.
For more information on U-turns,
see the tRNA structure
tutorial or the stereo picture
of tRNA U-turns.
IIb. Domain II
Fig. 4 of reference 1 shows the "domain II"
region of the hammerhead <
>. This region of conserved residues includes two G·A
base pairs, an unusual A-U pair and a standard G-C pair. To see
the h-bonds of the distal half of domain II click here
<>. To see the H-bonds of the proximal
half, click here <>.
There is also a short tutorial on a second
hammerhead ribozyme crystal structure available. You can also
see the two 3D structures
side-by-side or a red/blue
stereo picture of both structures.
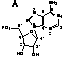 Nucleotide
numbering help.
Nucleotide
numbering help.
 tRNA
structure.
tRNA
structure.
 Hammerhead
Ribozyme - (Ref. 2).
Hammerhead
Ribozyme - (Ref. 2).
 Side-by-side
Comparison of Hammerhead Structures.
Side-by-side
Comparison of Hammerhead Structures.
 Red/blue
stereo picture of both Structures.
Red/blue
stereo picture of both Structures.
 Back
to intro to DNA-RNA structure.
Back
to intro to DNA-RNA structure.
Comments or Suggestions to:Jim
Nolan at jnolan@tulane.edu

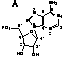



![]() Back
to intro to DNA-RNA structure.
Back
to intro to DNA-RNA structure.